In previous articles, I have surveyed various examples of recurring design logic in bacterial systems — in particular, two-component regulatory systems (here) and transcriptional hierarchies (here and here). In this article, I will discuss another example of recurring design logic — the control mechanisms of the expression of operons. Operons, you might recall, are clusters of genes that are transcribed together, under the control of a common promoter. There are, in fact, multiple ways in which operon control exhibits recurring design logic. Here, I will be focusing on similarities in repression mechanism between two evolutionarily unrelated operons — the tryptophan and arginine operons.
The Tryptophan Operon
A particularly well studied example of operon repression is the tryptophan (Trp) operon, which, together with the lactose operon, is often used to introduce the subject of operons to college students. The Trp operon is a sequence of five structural genes (TrpE, TrpD, TrpC, TrpB, and TrpA) that are transcribed together into one long mRNA transcript. The genes are separated from one another on the transcript by stop codons, and are translated into five proteins components that build three enzymes involved in the biosynthesis of tryptophan. The bacterial cell needs a mechanism for ensuring that these genes are only expressed when there are low levels of tryptophan available.
The Trp operon is represented by the figure below.
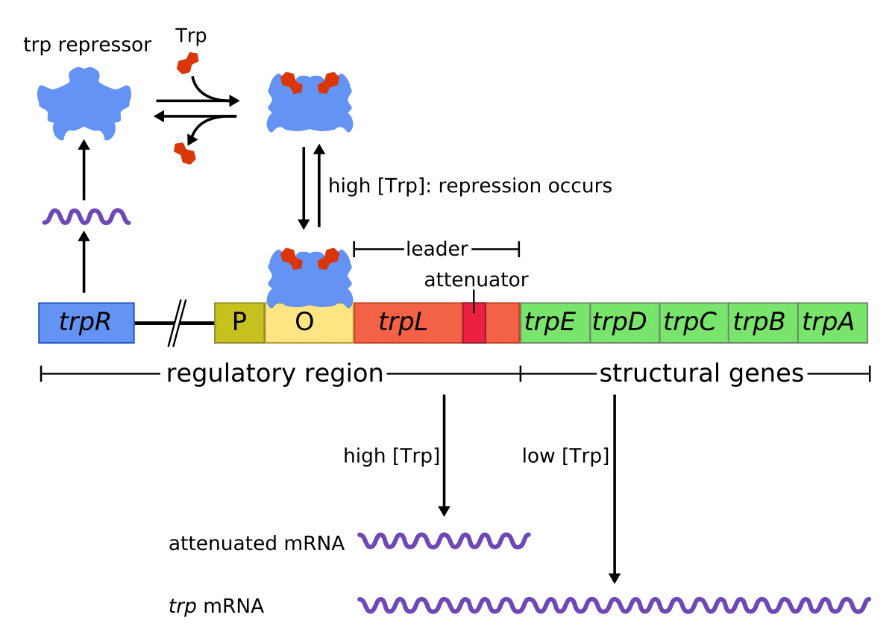
As shown in the diagram above, when tryptophan is abundant, it binds to the repressor. This causes a conformational change, facilitating the repressor’s binding to the operator sequence (labelled “O”), which is downstream of the promoter (labelled “P”). This blocks the RNA polymerase from transcribing the Trp operon.
The Arginine Operon
Compare the mechanism of controlling the Trp operon with the regulation of the arginine operon, illustrated below.
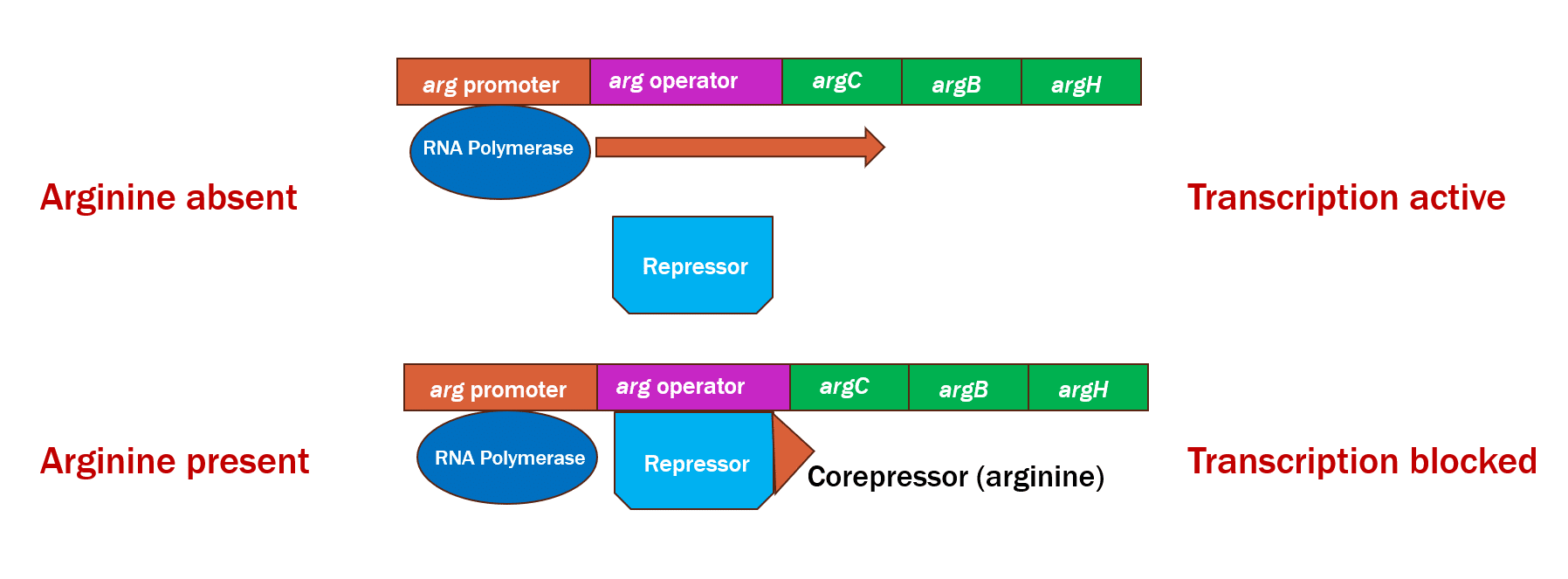
Just like the tryptophan operon, the repressor only docks to the operator when it is bound by arginine. By associating with the operator, the RNA polymerase is inhibited from transcribing the genes needed for the biosynthesis of arginine.
Recurring Design Logic
As we see in these two examples, the design logic is the same. And yet, these two systems are not evolutionarily related to one another — as evidenced by the fact that the enzymes encoded by these two operons do not share sequence homology, indicating distinct evolutionary origins. Moreover, the regulator proteins (TrpR and ArgR) are also structurally and genetically distinct, though both are members of the helix-turn-helix DNA binding protein superfamily (but this is a common structural motif). In a second article, I will discuss the recurring design logic exhibited by a second layer of regulation, known as attenuation.